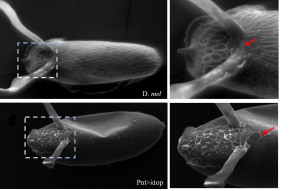
 My research focuses on screening DNA fragments that guide gene expression. Organs in our body develop from a single layer of epithelial cells. These cells are instructed to express different genes in sub-populations of cells (a process called tissue patterning) and consequently drive the formation of organs. I use egg development, oogenesis, in the fruit fly Drosophila melanogaster, as a model system to study how genes are expressed non-uniformly. Specifically, I focus on the patterning of follicle cells (FCs), a layer of epithelial cells that surround the developing oocyte. While FCs’ patterning has been vastly documented, our understanding of genes’ regulation is very limited. To find regulatory domains, I screened a large collection of fly lines that represent the dynamic expression of ~20 genes. Since gene regulation is a complex task, I have screened 230 unique fly lines. I was very excited with each new line that gave me a positive expression.
My research focuses on screening DNA fragments that guide gene expression. Organs in our body develop from a single layer of epithelial cells. These cells are instructed to express different genes in sub-populations of cells (a process called tissue patterning) and consequently drive the formation of organs. I use egg development, oogenesis, in the fruit fly Drosophila melanogaster, as a model system to study how genes are expressed non-uniformly. Specifically, I focus on the patterning of follicle cells (FCs), a layer of epithelial cells that surround the developing oocyte. While FCs’ patterning has been vastly documented, our understanding of genes’ regulation is very limited. To find regulatory domains, I screened a large collection of fly lines that represent the dynamic expression of ~20 genes. Since gene regulation is a complex task, I have screened 230 unique fly lines. I was very excited with each new line that gave me a positive expression.
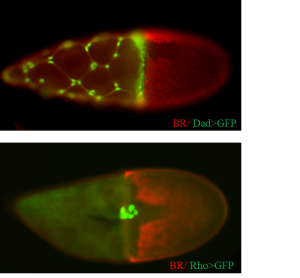
Overall, about 25% of the tested lines expressed the GFP reporter in the follicle cells. I have been very happy to discover that about 25% of the positive fly lines showed a partial or full expression pattern of the corresponding endogenous genes. I am currently using a new technique, RNAseq, to localize the regulatory information of genes to certain positions in the genes’ locus. Finally, the flies that I found in my screen were already used by me and other students in the Yakoby Lab to mutate genes in sub-populations of epithelial cells.
